[ Source: Juan Siliezar | The Harvard Gazette | July 07, 2021 ]
Microscopists have long sought a way to produce high-quality, deep-tissue imaging of living subjects in a timely fashion. To date, they have had to choose between image quality or speed when looking into the inner workings of complex biological systems.
A better imaging system would have a powerful impact on researchers in biology and in neuroscience, experts say. Now Dushan N. Wadduwage, a John Harvard Distinguished Science Fellow in imaging at the FAS Center of Advanced Imaging, along with a team from MIT, has detailed a new technique that would make that possible in a report in Science Advances.
In the paper, the team presents a new process that uses computational imaging to get high-resolution images 100 to 1,000 times faster than other state-of-the-art technologies using complex algorithms and machine learning. The method can shorten a process that often takes months into a matter of days.
The system, called De-scattering with Excitation Patterning (or DEEP), is believed to be the first of its kind and may one day lead to new understanding of how complicated processes, such as those in the brain, function, because DEEP can capture images other microscopes cannot.
Because the new system has the potential to actually speed up what they can image along with how fast they can do it, “scientists will be able to image fast processes they haven’t been able to capture before, like what happens when a neuron fires or how the signals move around in the brain,” Wadduwage said. “Also, because it’s technically faster, you can image a larger volume of area at one time, not just a small field of view as you would with a slower imaging system. It’s like being able to look at a much larger picture, and this is very important for neuroscientists and other biologists to actually get better statistics as well as to see what’s happening around the area being imaged.”
The system works like many other animal-imaging techniques. Near-infrared laser light is used to penetrate deep through biological tissue that scatters the light. That light excites the fluorescent molecules the researchers want to image, and these emit signals that the microscope captures to form an image.
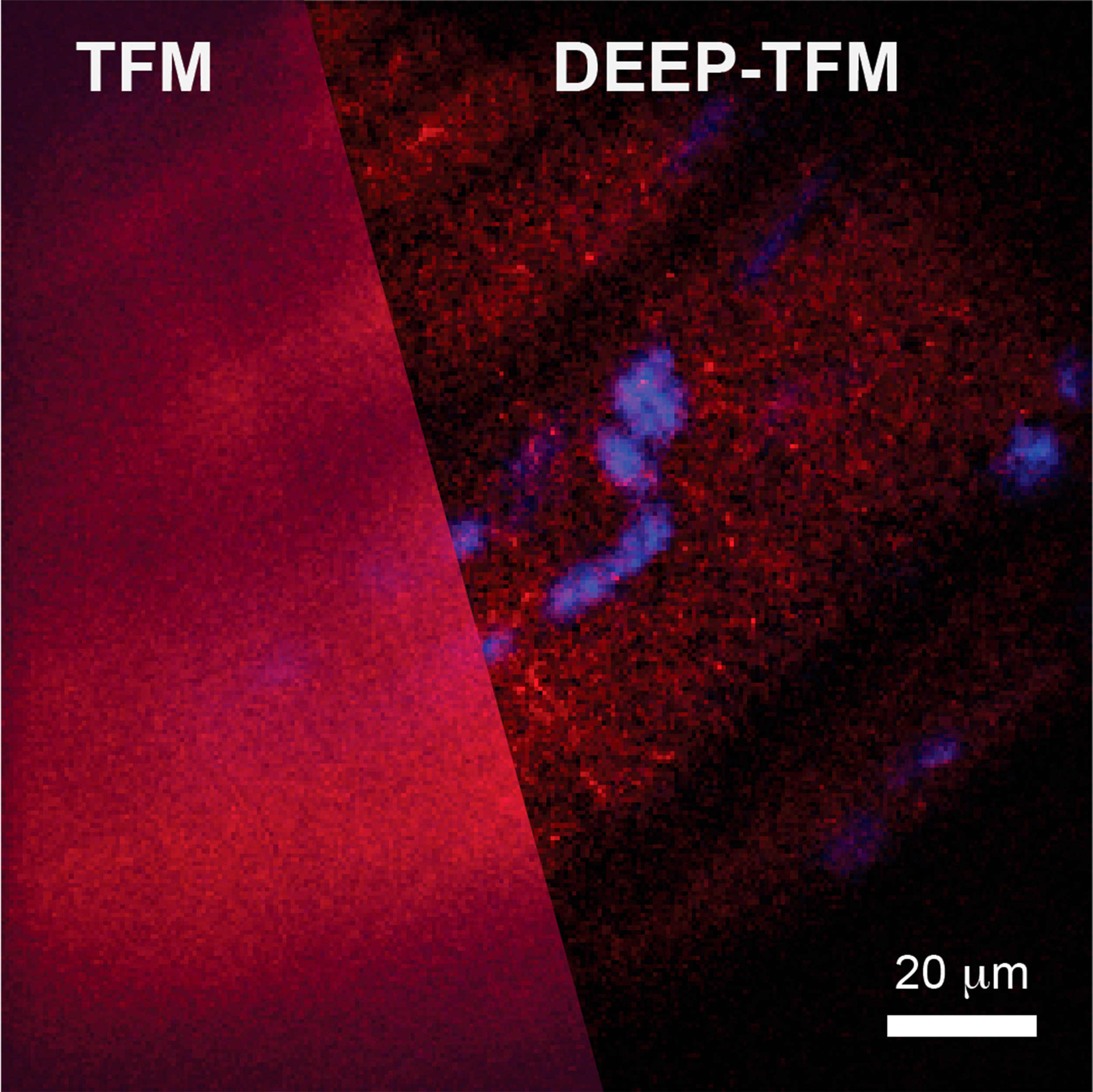
There have been two main ways these types of images are taken. Point-scanning multiphoton microscopy can penetrate deep into a specimen and capture high-quality images, but this process is extremely slow because the image is formed one point at a time. Capturing a centimeter-sized image, for example, can take months. It also limits studies of fast biological dynamics, such as neurons firing. The other method is called temporal focusing microscopy. This is much faster and can capture images at a wider scale, but it is unable to capture high-resolution images at anything deeper than a few millionths of a meter. The fluorescent light scatters too much, causing the image to degrade when the camera detects it.
DEEP-TFM imaging of cortical vasculature at 100 um below the surface. DEEP-TFM illumination is shown in the red channel in the left panel; acquired images are shown in the green channel in the left panel; the reconstruction is shown in the right panel. The reconstruction improves with the increasing number of illumination patterns.
DEEP, however, allows for wide-scale and quick tissue penetration, and produces high-resolution images. The system projects a wide light into the subject as in the temporal microscopy method, but that laser light is in a specific pattern. The computational-imaging algorithm that knows the initial pattern takes in the information gathered to reverse the process when it gets scattered and then reconstructs it, de-scattering the image. This is especially notable since it takes the reconstruction of structural features from millions of measurements to tens and hundreds. DEEP can image hundreds of microns deep through scattering tissue comparable to point-scanning techniques.
