[ Source: Scientifica ]
Murat Yildirim, Picower Institute for Learning and Memory
Deep tissue imaging has been my great passion since my doctoral studies. During my PhD, I developed two- and three-photon systems to perform deep tissue imaging in ex vivo tissue samples as well as precise tissue ablation. Then, I realised the possible applications of two- and three-photon microscopy in brain imaging, thus I began a joint collaborative project with Mriganka Sur and Peter So at MIT. Since then, I have been learning to ask fundamental questions in neuroscience and developing multiphoton systems to find the answers to these questions.
How was Scientifica’s SliceScope used to build the three-photon system?
When I started to design my scope, one of the first things that I was searching for was a translational stage for moving the sample in lateral and axial directions. Since we knew we did not want to move the animal, especially in the axial direction, I needed two separate stages for lateral and axial movements. Another important criterion in stage selection was not having any electrical noise appearing in images acquired by the photomultipier tubes (PMTs).
For the noise issue, I consulted a couple of my colleagues who have performed various electrophysiology experiments with slices in our lab and they recommended I use Scientifica stages. I purchased Scientifica’s translational stages and SliceScope, and designed the rest of my system around these stages. Since the Slicescope has enough space in the axial direction, I could design my system both for small and big animal experiments.
What has been investigated using the system?
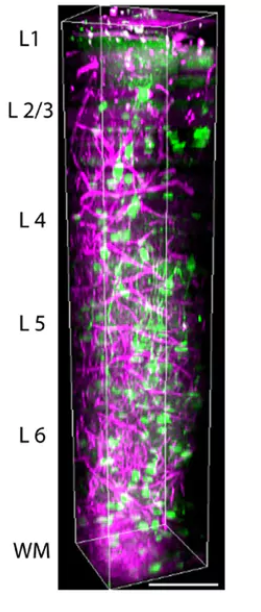
Developing a custom-made three-photon microscope requires optimising laser parameters such as repetition rate and pulse width as well as microscope parameters such as aberration, excitation and emission path optics. By optimising these parameters, we believe that we can make novel discoveries that were not possible before.
Select Media Coverage: